Our Research
Our lab combines computational methods with protein characterization. Computational methods include superposition, phylogenetics, and ancestral sequence reconstruction. The wet lab components mostly involve expression and characterization of modern and ancestral proteins to determine how their characteristics have changed over evolutionary time.
Recent Publications
-
-
-
-
-
A gustatory receptor paralog controls rapid warmth avoidance in Drosophila
Likelihood-based classification of cryo-EM images using FREALIGN
BAYESIAN ALIGNMENT OF SIMILARITY SHAPES
Likelihood and Empirical Bayes Superposition of Multiple Macromolecular Structures
Potassium-selective block of barium permeation through single KcsA channels
-
How has enzyme function, structure, and mechanism evolved?
Despite the fundamental biological importance of enzyme function, there are still many basic unresolved problems in molecular evolution, each of which has important implications for understanding how modern enzymes function and how to rationally design novel activities.
For example: 1. How have complicated enzyme kinetic mechanisms evolved? 2. Are ancestral enzymes functionally promiscuous? 3. How do substitutions distant from the enzyme active site affect activity? 4. How many ways can an enzyme perform the same catalytic function (convergence)?
How good are ancestral reconstructions?
We developed a cross-validation method called Extant Sequence Reconstruction (ESR) that allows to statistically test ASR methodology. Click here to learn more about how we are using ESR for model and alignment selection..
How do metalloenzymes evolve novel catalytic mechanisms?
How have homologous metalloenzymes evolved to convert similar substrates into vastly different products? AetD and SktA are both members of the heme oxygenase-like diiron oxygenase (HDO) superfamily and exhibit striking structural similarity. While both proteins use halogenated L-Trp as their substrates, AetD creates an indole-carboxynitrile moiety and glycolic acid and SktA creates skatole and cyanide. In a collaboration with Maria Pandelia, we are using Ancestral Sequence Reconstruction and Ancestral Protein Resurrection to identify key events along AetD’s and SktA’s evolutionary history that may define their divergent chemistries.
What is the evolutionary trajectory of enzyme mechanisms?
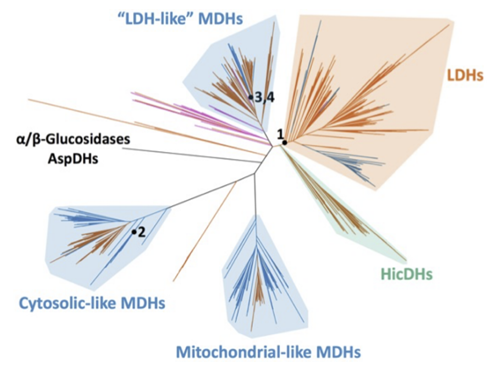
There is a lack of research specifically tracing the evolutionary trajectory of enzymes transitioning between ordered and random sequential binding mechanism. Previously, some have hypothesized that ordered binding evolved first, but this has not yet been supported experimentally. We are investigating the evolutionary development of enzymatic mechanism by looking at both extant and ancestral Lactate and Malate Dehydrogenase enzymes from the apicomplexan phylum.
Bi-specificity
How biophysically accurate are ancestrally resurected proteins?
In common practice, the single most probable (SMP) ancestral sequence is reconstructed to choose the most probable amino acid at every site of the ancestral protein. Despite the tangible success of ASR, the accuracy of its reconstructions is currently unknown. Currently there is a controversy regarding the apparent increased thermostability of proteins resurrected using ASR methodology: is increased thermostability a property of ancestral proteins, a result of sampling bias, or a systematic artifact of the methodology used? A better alternative for estimating is to sample from the ancestral distribution. An on going project in our lab is aimed at more thoroughly sampling from the distribution, and assessing how methodology impacts biophysical parameters beyond thermostability.